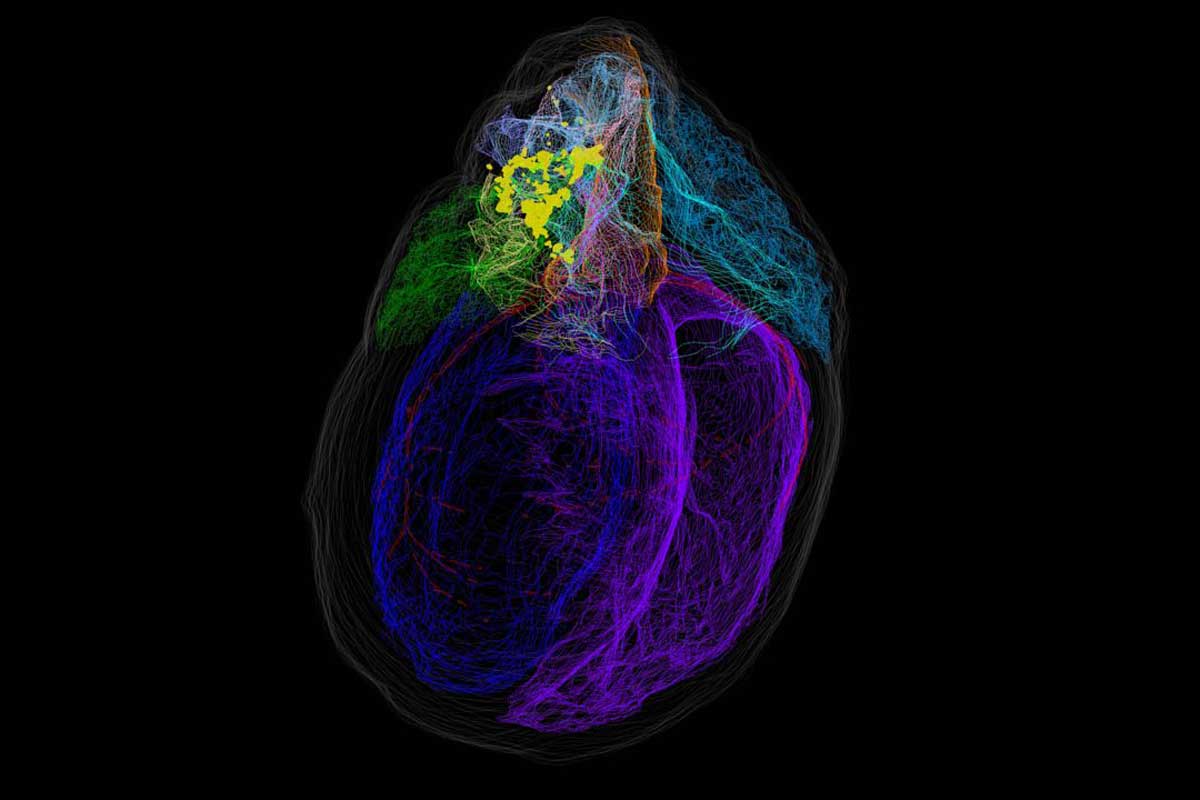
A team of UCF College of Medicine scientists has helped develop a virtual 3D heart that shows the body’s neurons and their connection to the heart — a tool they hope will help develop better life-saving therapies for cardiac disease. The researchers say this is the first model that has been created that digitally showcases the heart’s unique network of neurons.
The study published in Cell Press’ iScience journal, was the project of an interdisciplinary team of researchers from UCF, Thomas Jefferson University and industry partners Strateos and MBF Bioscience.
Principal investigator of the UCF team, neuro-cardiovascular scientist Zixi Jack Cheng, said the project mapped thousands of neurons in clusters around the heart that help to control heartbeat and blood circulation.
“This project was geared at creating a blueprint of how the body’s intrinsic cardiac nervous system is anatomically connected to the heart structure, by mapping the pathways of these neurons and identifying their functions as it relates to how they control the heart,” Cheng says.
Over a two-year period, the collaborators created a comprehensive map of the cardiac nervous system at the cellular level, mapping specific neuron clusters called ganglia. The researchers say this blueprint will allow scientists and physicians to more precisely study the neuroanatomy of the heart.
“With the spatial and the gene-expression mapping of the intrinsic cardiac nervous system, we can begin to discuss the precise roles that these neurons play,” says Cheng. “Do separate clusters of neurons have different functions, or do they work into an integrated way to influence heart health? Now we can address these questions in a way that wasn’t possible before.”
The map will also have important clinical applications for treating patients with heart disease.
The map will also have important clinical applications for treating patients with heart disease. By understanding a patient’s anatomical structures and functions, healthcare providers may be able to manipulate specific nerves to emit bioelectrical signals that repair the heart and effectively treat heart conditions.
Cheng’s research assistant and co-author of the study, Clara Leung, explained that the mapping may be helpful in treating atrial fibrillation, or irregular heartbeat.
“There are specific neurons that are concentrated around the pulmonary veins and on the atria that are responsible for sending signals to the sinoatrial node in the heart,” says Leung, who worked on the project for her master’s thesis. “And so, by identifying specific clusters in these areas and manipulating them, we may eventually be able to control some of these abhorrent signaling that are causing abnormal contractions in the heart.”
Jin Chen, a postdoctoral fellow in Cheng’s lab and a co-author of the study, is now injecting colored tracers into the heart of animal models to map the pathways neurons use as they send and receive signals from the cardiac nervous system. The research team hopes this will help them further develop a digital, interactive 3D map to better understand heart disease.
“So far we have worked on normal animal models, but it’s also really important to compare the differences between healthy and diseased models down the road, because that’s our end goal for the study – to help patients with abnormalities,” she says.
The project was funded by the National Institutes of Health under the Stimulating Peripheral Activity to Relieve Conditions program and drew on technologies and expertise from multiple research groups. SPARC supports research into understanding nerve-to-organ interactions with the aim of developing bioelectronic treatments for diseases and conditions.
Analysts estimate that approximately every 40 seconds an American will have a heart attack. Cardiovascular disease is the No. 1 cause of death in the U.S. and globally, accounting for an estimated 18 million deaths each year according to the World Health Organization.
The digital map of the heart is available on the SPARC’s online portal, allowing other research teams to access the data.